dataset_name: CROssBARv2-KG
tags:
- biomedical
- knowledge-graph
- bioinformatics
- biology
- chemistry
- systems-biology
task_categories:
- graph-construction
- link-prediction
- node-classification
- graph-ml
pretty_name: CROssBARv2 Knowledge Graph Dataset
language:
- en
size_categories:
- 10M<n<100M
CROssBARv2-KG
This repository provides the dataset for the CROssBARv2 Knowledge Graph (KG), a heterogeneous and general-purpose biomedical KG-based system.
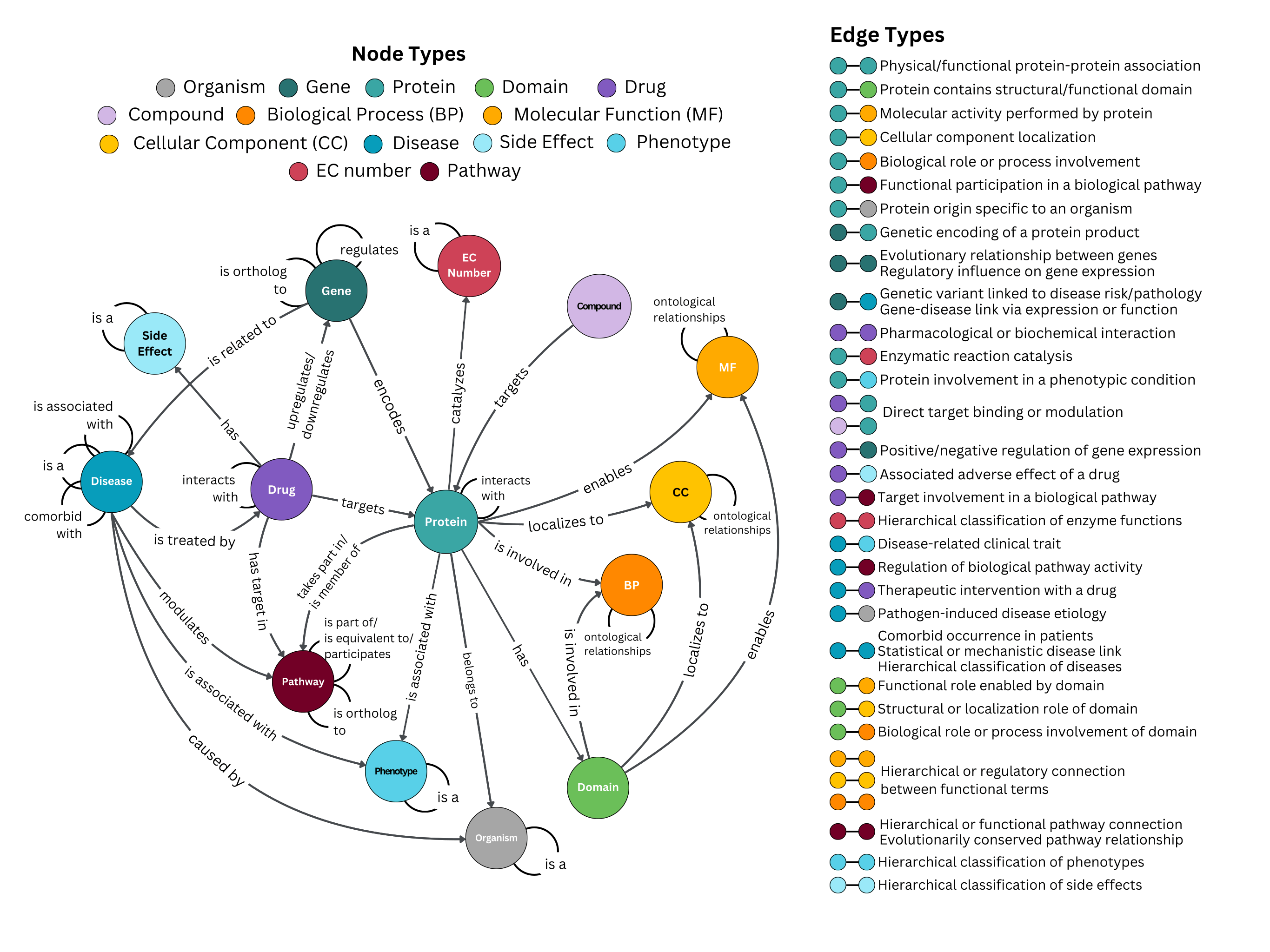
The CROssBARv2 KG comprises approximately 2.7 million nodes spanning 14 distinct node types and around 12.6 million edges representing 51 different edge types, all integrated from 34 biological data sources. We also incorporated several ontologies (e.g., Gene Ontology, Mondo Disease Ontology) along with rich metadata captured as node and edge properties.
Building upon this foundation, we further enhanced the semantic depth of CROssBARv2. This was achieved by generating and storing embeddings for key biological entities, such as proteins, drugs, and Gene Ontology terms. These embeddings are managed using the native vector index feature in Neo4j, enabling powerful semantic similarity searches.
Data
The dataset is organized into two primary directories:
nodes/: Contains CSV files defining the biological entities.edges/: Contains CSV files defining the relationships between these entities.
nodes
Each file corresponds to a specific biological entity type and contains a primary identifier column that provides a unique ID along with additional metadata fields. Identifiers follow the Compact URI (CURIE) standard (e.g., uniprot:Q9H161) as defined in the Bioregistry.
The table below lists the primary identifier column name for each node file:
| File | ID Column Name |
|---|---|
| Cellular_component.csv | id |
| Biological_process.csv | id |
| Molecular_function.csv | id |
| Protein.csv | id |
| Phenotype.csv | hpo_id |
| Pathway.csv | pathway_id |
| Ec.csv | ec_number |
| Compound.csv | compound_id |
| Gene.csv | id |
| Side_effect.csv | meddra_id |
| Drug.csv | drugbank_id |
| Drug.csv | drugbank_id |
| Organism.csv | id |
| Domain.csv | id |
| Disease.csv | disease_id |
The following table demonstrates the CURIE format used for each node type within the KG:
| Node Type | CURIE |
|---|---|
| Protein | uniprot:Q9H161 |
| Gene | ncbigene:60529 |
| OrganismTaxon | ncbitaxon:9606 |
| ProteinDomain | interpro:IPR000001 |
| Drug | drugbank:DB00821 |
| Compound | chembl:CHEMBL6228 |
| GOTerm (BiologicalProcess, MolecularFunction, CellularComponent) | go:0016072 |
| Disease | mondo:0054666 |
| Phenotype | hp:0000012 |
| SideEffect | meddra:10073487 |
| EcNumber | eccode:1.1.1.- |
edges
Each file represents a relationship between two biological entity types in the KG.
Every edge file contains source and target columns, which specify the identifiers of the nodes being linked.
Other columns serve as metadata for the relationship.
The table below lists the filename along with the source and target identifier column names and a description of the edge type:
| File | Source Column | Target Column | Description |
|---|---|---|---|
| Pathway_orthology.csv | pathway1_id | pathway2_id | Evolutionarily conserved pathway relationship |
| Go_to_go.csv | source | target | Hierarchical or regulatory connection between functional terms |
| Gene_to_disease_edge.csv | gene_id | disease_id | Gene-disease link via expression or function |
| Ec_hierarchy.csv | child_id | parent_id | Hierarchical classification of enzyme functions |
| Disease_to_drug_edge.csv | disease_id | drug_id | Therapeutic intervention with a drug |
| DTI.csv | drugbank_id | uniprot_id | Direct target binding or modulation |
| Phenotype_hierarchical_edges.csv | child_id | parent_id | Hierarchical classification of phenotypes |
| Protein_to_phenotype.csv | protein_id | hpo_id | Protein involvement in a phenotypic condition |
| Drug_to_side_effect.csv | drugbank_id | meddra_id | Associated adverse effect of a drug |
| Disease_to_disease_comorbidity_edge.csv | disease1 | disease2 | Comorbid occurrence in patients |
| DGI.csv | entrez_id | drugbank_id | Positive/negative regulation of gene expression |
| Drug_to_pathway.csv | drug_id | pathway_id | Target involvement in a biological pathway |
| PPI.csv | uniprot_a | uniprot_b | Physical/functional protein-protein association |
| Disease_to_disease_association_edge.csv | disease_id1 | disease_id2 | Statistical or mechanistic disease link |
| Phenotype_to_disease.csv | hpo_id | disease_id | Disease-related clinical trait |
| Orthology.csv | entrez_a | entrez_b | Evolutionary relationship between genes |
| Side_effect_hierarchy.csv | child_id | parent_id | Hierarchical classification of side effects |
| Organism_to_disease_edge.csv | organism_id | disease_id | Pathogen-induced disease etiology |
| DDI.csv | drug1 | drug2 | Pharmacological or biochemical interaction |
| Protein_to_ec.csv | protein_id | ec_id | Enzymatic reaction catalysis |
| Reactome_hierarchical_edges.csv | child_id | parent_id | Hierarchical or functional pathway connection |
| Tf_gene_edges.csv | tf | target | Regulatory influence on gene expression |
| Disease_hiererchical_edges.csv | child_id | parent_id | Hierarchical classification of diseases |
| Protein_to_pathway.csv | uniprot_id | pathway_id | Functional participation in a biological pathway |
| Protein_has_domain.csv | source_id | target_id | Protein contains structural/functional domain |
| Protein_belongs_to_organism.csv | source_id | target_id | Protein origin specific to an organism |
| Protein_to_go.csv | source | target | Molecular activity performed by protein / Cellular component localization / Biological role or process involvement |
| Pathway_to_pathway.csv | pathway_id1 | pathway_id2 | Hierarchical or functional pathway connection |
| Gene_encodes_protein.csv | source_id | target_id | Genetic encoding of a protein product |
| Disease_to_pathway.csv | disease_id | pathway_id | Regulation of biological pathway activity |
| Domain_to_go.csv | source | target | Functional role enabled by domain / Structural or localization role of domain / Biological role or process involvement of domain |
| CTI.csv | chembl | uniprot_id | Direct target binding or modulation |
How to Use
You can easily load this dataset using the Hugging Face datasets library:
from datasets import load_dataset
# Example 1: Load the Protein nodes
proteins = load_dataset("HUBioDataLab/CROssBARv2-KG", data_files="nodes/Protein.csv")
# Example 2: Load Drug-Target Interactions (Edges)
dti_edges = load_dataset("HUBioDataLab/CROssBARv2-KG", data_files="edges/DTI.csv")